Article By : Abhay Chaturvedi
Much of the investigations of the classical genetics have been devoted to the localization of genes on chromosomes. As according to chromosome mapping, gene mapping depends on the assumption that genes do not move from one position to another. To a great extent, the hypothesis has been found correct. In fact most genes occupy fixed sites on chromosome and the overall structure of the genetic map is practically invariant. However in early 1940s researchers have found that some DNA sequences can actually change their position.
These mobile elements have been variously called Jumping genes, Mobile elements, Transposons etc. The formal name is Transposable elements and their movement is called transposition. The term transposon was coined by R.W. Hedges and A.E. Jacob in 1974.
Definition
Transposable elements can be defined as small mobile DNA sequence that move around chromosome with no regard for Haemology and insertion of these elements may produce deletions inversions chromosomal fusions and even more complicated rearrangements.
Discovery of Transposable Elements
Transposable elements were discovered by Barbara McClintock in 1940s through an analysis of genetic inability in maize (corn). The instability involved chromosome breakage and was found to occur at sites where transposable elements were located. In McClintock’s analysis, the events of breakage were detected by following the loss of certain genetic marker.
In some experiments, McClintock used a marker (gene) that controlled the deposition of pigmentation in the aleurone which is the outer most layer of endosperm of maize kernels. McClintock’s marker was an allele of the locus C on the short arm of an chromosome I. Since this allele is dominant inhibitor of aleurone colouration any kernel possessing it should be colourless. He fertilized CC ears with pollen from C1C1 tessels producing kernels in which the endosperm was C1CC. Although many of kernels were colourless, as except some showed patches of brownish-purple pigment. McClintock guessed that in such mosaics, the inhibitory ‘C’ allele had been lost sometime during endosperm development leading to a clone of tissue that was capable of producing pigment. The genotype in such a clone should – CC, where the clash indicates loss of C1 allele.
Characters of Transposable Elements
- They were found to be DNA sequences that code for enzymes which bring about the insertion of an identical copy of themselves into a new DNA site.
- Transposition events involve both recombination and replication processes which frequently generate two daughter copies of the original transposable elements. One copy remains at the parent site while the other appears at the target site (on the host chromosome).
- The insertion of transposable elements invariably disrupts the integrity of their target genes. For example, if an IS (= insertion sequence) becomes inserted into an Operon, it interrupts the coding sequence and inactivates the expression of the target gene into which it inserts as well as any gene downstream in that same Operon. This is because an IS contains transcription and/or translation termination signals that block the expression of other genes downstream in an Operon. This “one-way” mutational effect (or polarity) is referred to as a polar mutation.
- Since transposable elements carry signals for the initiation of RNA synthesis, they sometimes activate previously dormant genes.
- A transposable element is not a replicon (a sequence that contains a site for the origin of replication), thus, it cannot replicate apart from the host chromosome the way that plasmids and phage can.
- No homology exists between the transposon and the target site for its insertion. Many transposons can insert at virtually any position in the host chromosome or into a plasmid. Some transposons seem to be more likely to insert at certain positions (hot spots), but rarely at base- specific target sites.
Types of Transposable Elements :
Transposable elements are of following types
Insertion sequence (IS) or Transposon (simple)
The insertion sequence are shorter and do not code for protein. In fact it’s carry the genetic information necessary for their transposition (ie, the gene for enzyme transposes)
Transposon (Tn) or complex Transposon
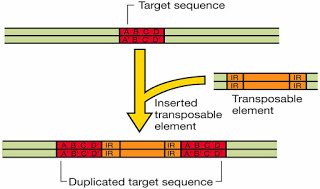
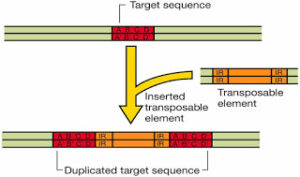
These are several thousand base pairs long and have gene coding for one or more protein (including resistance factors in bacteria which act against antibiotics). The hallmark of a tranposon is the presence of identical inverted terminal repeat (IR) sequence of 8 to 38 base pairs. Each type of transposon has its own unique inverted repeat. On either side of a transposon is a short (less than 10 bp) direct repeat. If the transposon exists in multiple copies, these direct repeats are of different base composition at each site. where the transposon exists in the chromosome; the inverted terminal repeats, however remain the same for a given tranposon. The sequence into which a transposable element insert is called the target sequence.
During insertion of a transposon, the singular target sequence becomes duplication, and thus appears as direct repeats the flanking of inserted transposable element. The direct repeats are not considered part of the transposon. These repeat sequences themselves act like IS or Is like segments.
Examples of Transposon
- Tn 3 transposon of E. coli:
The molecular structure of transposon Tn 3 of E. coli has been worked out (Fig. 41.4). Tn 3 has 4957 bp and contains three genes such as tnp A, tnp R and bla, coding respectively for the following proteins: 1. transposase having 1015 amino acids and required for transposition; 2. a repressor (also called resolvase) containing 185 amino acids which regulates the transposase; and 3. β- Lactamase enzyme which confers resistance to the antibiotic ampicillin. On both sides of the Tn 3 is an inverted repeat of about 38 bp.
The bacteriophage Mu (.Mu = mutator) is a temperate bacteriophage having typical phage properties and could be regarded as a giant transposon. Phage Mu was thought to behave in a unique manner since it does not multiply in a lytic way. It always starts its life-cycle by inserting itself into the E. coli chromosome at random locations. The resulting prophage often mutates the genes into which they become inserted. Thus, phage Mu was originally known for its mutagenic properties. During later part of phage life cycle, upon receipt of an appropriate signal, the Mu prophage somehow generates its transposition, but also activates the genes encoding for structural proteins which package its DNA. Mu phage is unique also in transposing much often than other transposons, providing nearly a hundred new copies of Mu during its hour-long life-cycle.
Like other transposons, Mu phage has inverted repeats (IR) at or near the ends of its DNA and it makes a short duplication with 5 base pairs of the bacterial DNA into which it is inserted.
The yeast Saccharomyces cerevisiae carries about 35 copies of a transposable element called Ty in its haploid genome. These transposons are about 5900 base pairs long and are bounded at each end by a DNA segment called the delta (8) sequence, which is approximately 340 base-pairs long.
Each 8 sequence is oriented in the same direction, forming what are known as direct long terminal repeats or LTRs. Ty elements are flanked by five base-pair direct repeats created by the duplication of DNA at the site of the Ty insertion. These target site duplications are found rich in A-T base pairs. Yeast Ty elements are sometimes called retrotransposons because of their overall similarity to the retroviruses. Like the yeast, in other eukaryotes such as Zea Mays, Drosophila, mice and snapdragons (Antirrhinum majus) transposable elements of one or other type are found.